Tetratricopeptide
| Tetratricopeptide repeat | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 |
|||||||||
| Identifiers | |||||||||
| Symbol | TPR_1 | ||||||||
| Pfam | PF00515 | ||||||||
| Pfam clan | CL0020 | ||||||||
| InterPro | IPR001440 | ||||||||
| SCOP | 1a17 | ||||||||
| SUPERFAMILY | 1a17 | ||||||||
| CDD | cd00189 | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
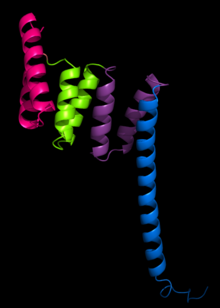
The tetratricopeptide repeat (TPR) is a structural motif. It consists of a degenerate 34 amino acid sequence motif identified in a wide variety of proteins. It is found in tandem arrays of 3–16 motifs, which form scaffolds to mediate protein–protein interactions and often the assembly of multiprotein complexes. These alpha-helix pair repeats usually fold together to produce a single, linear solenoid domain called a TPR domain. Proteins with such domains include the anaphase-promoting complex (APC) subunits cdc16, cdc23 and cdc27, the NADPH oxidase subunit p67-phox, hsp90-binding immunophilins, transcription factors, the PKR protein kinase inhibitor, the major receptor for peroxisomal matrix protein import PEX5 and mitochondrial import proteins.
The structure of the PP5 protein was the first structure to be determined. The structure solved by X-ray crystallography by Das and colleagues showed that the TPR sequence motif was composed of a pair of antiparallel alpha helices. The PP5 structure contained 3 tandem TPR repeats which showed the sequential TPR repeats formed an alpha-helical solenoid structure.
A typical TPR structure is characterized by interactions between helices A and B of the first motif and helix A’ of the next TPR. Although the nature of such interactions may vary, the first two helices of the TPR motif typically have a packing angle of ~24 degrees within a single motif. Repeats of more than three TPR motifs generate a right handed superhelix characterized by both a concave and a convex face, of which the concave face is usually involved in ligand binding.
...
Wikipedia
