Serine hydroxymethyltransferase
| Serine Hydroxymethyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
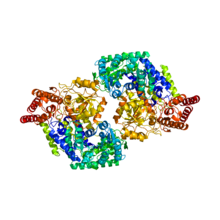
PyMol rendered crystal structure of serine hydroxymethyltransferase
|
|||||||||
| Identifiers | |||||||||
| EC number | 2.1.2.1 | ||||||||
| CAS number | 9029-83-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
|
|||||||||
| Search | |
|---|---|
| PMC | articles |
| PubMed | articles |
| NCBI | proteins |
Serine hydroxymethyltransferase (SHMT) is a Pyridoxal phosphate (PLP) (Vitamin B6) dependent enzyme (EC 2.1.2.1) which plays an important role in cellular one-carbon pathways by catalyzing the reversible, simultaneous conversions of L-serine to glycine and tetrahydrofolate (THF) to 5,10-methylenetetrahydrofolate (5,10-CH2-THF). This reaction provides the largest part of the one-carbon units available to the cell.
The structure of the SHMT monomer is remarkably similar across prokaryotes and eukaryotes, but whereas the active enzyme is a dimer in prokaryotes, the enzyme exists as a tetramer in eukaryotic cells, though the evolutionary basis for this difference in structure is unknown. However, the evolutionary path taken by SHMT going from prokaryotic dimeric form to the eukaryotic tetrameric form can be easily seen as a sort of doubling event. In other words, the eukaryotic SHMT tetramer resembles two prokaryotic dimers that have packed together, forming what has been described as a “dimer of dimers.” The interaction between two monomers within a dimer subunit has been found to occur over a greater contact area and is thus much tighter than the interaction between the two dimers.
A single SHMT monomer can be understood in terms of three domains: an N-terminus “arm,” a “large” domain, and a “small” domain. The N-terminus arm appears to be critical for the maintenance of the tight interaction between two monomers. The arm, consisting of two alpha helices and a beta sheet, wraps around the other monomer when in oligomeric form. The “large” domain contains the PLP binding site, which resembles the PLP binding domains of related PLP dependent proteins, like aspartate aminotransferase. The large domain in the eukaryotic form also contains a histidine that has been found essential for tetramer stability. All four histidines of these residues, one from each monomer, sit at the center of the tetrameric complex, where two histidines from a dimeric subunit engages in stacking interactions with the histidines of the other subunit. Prokaryotic SHMT has a proline residue rather than histidine in the equivalent position, which would in part explain why prokaryotic SHMT does not form tetramers.
...
Wikipedia
