Ubiquitination
| Ubiquitin family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
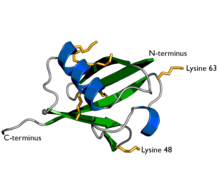
A diagram of ubiquitin. The seven lysine sidechains are shown in yellow/orange.
|
|||||||||
| Identifiers | |||||||||
| Symbol | ubiquitin | ||||||||
| Pfam | PF00240 | ||||||||
| InterPro | IPR000626 | ||||||||
| PROSITE | PDOC00271 | ||||||||
| SCOP | 1aar | ||||||||
| SUPERFAMILY | 1aar | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
Ubiquitin is a small (8.5 kDa) regulatory protein that has been found in almost all tissues () of eukaryotic organisms. It was discovered in 1975 in Israel by Gideon Goldstein and further characterized throughout the 1970s and 1980s. There are four genes in the human genome that produce ubiquitin: UBB, UBC, UBA52 and RPS27A.
The addition of ubiquitin to a substrate protein is called ubiquitination or ubiquitylation. Ubiquitination can affect proteins in many ways: it can signal for their degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitination is carried out in three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade binds ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cysteine residues through a thioester bond, serine and threonine residues through an ester bond, or the amino group of the protein's N-terminus via a peptide bond.
...
Wikipedia
