UDP-glucose 4-epimerase
| UDP-glucose 4-epimerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
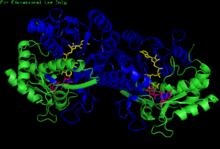
H. sapiens UDP-glucose 4-epimerase homodimer bound to NADH and UDP-glucose. Domains: N-terminal and C-terminal.
|
|||||||||
| Identifiers | |||||||||
| EC number | 5.1.3.2 | ||||||||
| CAS number | 9032-89-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
|
|||||||||
| Search | |
|---|---|
| PMC | articles |
| PubMed | articles |
| NCBI | proteins |
| UDP-galactose-4-epimerase | |
|---|---|

Human GALE bound to NAD+ and UDP-GlcNAc, with N- and C-terminal domains highlighted. Asn 207 contorts to accommodate UDP-GlcNAc within the active site.
|
|
| Identifiers | |
| Symbol | GALE |
| Entrez | 2582 |
| HUGO | 4116 |
| OMIM | 606953 |
| RefSeq | NM_000403 |
| UniProt | Q14376 |
| Other data | |
| EC number | 5.1.3.2 |
| Locus | Chr. 1 p36-p35 |
The enzyme UDP-glucose 4-epimerase (EC 5.1.3.2), also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.
Additionally, human and some bacterial GALE isoforms reversibly catalyze the formation of UDP-N-acetylgalactosamine (UDP-GalNAc) from UDP-N-acetylglucosamine (UDP-GlcNAc) in the presence of NAD+, an initial step in glycoprotein or glycolipid synthesis.
Dr. Luis Leloir deduced the role of GALE in galactose metabolism during his tenure at the Instituto de Investigaciones Bioquímicas del Fundación Campomar, initially terming the enzyme waldenase. Dr. Leloir was awarded the 1970 Nobel Prize in Chemistry for his discovery of sugar nucleotides and their role in the biosynthesis of carbohydrates.
GALE belongs to the short-chain dehydrogenase/reductase (SDR) superfamily of proteins. This family is characterized by a conserved Tyr-X-X-X-Lys motif necessary for enzymatic activity; one or more Rossmann fold scaffolds; and the ability to bind NAD+.
GALE structure has been resolved for a number of species, including E. coli and humans. GALE exists as a homodimer in various species.
While subunit size varies from 68 amino acids (Enterococcus faecalis) to 564 amino acids (Rhodococcus jostii), a majority of GALE subunits cluster near 330 amino acids in length. Each subunit contains two distinct domains. An N-terminal domain contains a 7-stranded parallel β-pleated sheet flanked by α-helices. Paired Rossmann folds within this domain allow GALE to tightly bind one NAD+ cofactor per subunit. A 6-stranded β-sheet and 5 α-helices comprise GALE's C-terminal domain. C-terminal residues bind UDP, such that the subunit is responsible for correctly positioning UDP-glucose or UDP-galactose for catalysis.
...
Wikipedia
