Protease inhibitor (biology)
| Peptidase inhibitor I9 |
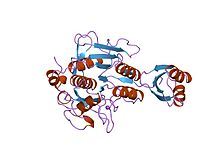
subtilisin bpn' prosegment (77 residues) complexed with a mutant subtilisin bpn' (266 residues). crystal ph 4.6. crystallization temperature 20 c diffraction temperature-160 c
|
| Identifiers |
| Symbol |
Inhibitor_I9 |
| Pfam |
PF05922 |
| InterPro |
IPR010259 |
| MEROPS |
I9 |
| SCOP |
1gns |
| SUPERFAMILY |
1gns |
|
|
| Serine endopeptidase inhibitors |

solution structure of marinostatin, a protease inhibitor, containing two ester linkages
|
| Identifiers |
| Symbol |
Inhibitor_I10 |
| Pfam |
PF12559 |
| InterPro |
IPR022217 |
|
|
| Cathepsin propeptide inhibitor domain (I29) |

crystal structure of a cysteine protease proform
|
| Identifiers |
| Symbol |
Inhibitor_I29 |
| Pfam |
PF08246 |
| InterPro |
IPR013201 |
|
|
In biology and biochemistry, protease inhibitors are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
In medicine, protease inhibitor is often used interchangeably with alpha 1-antitrypsin (A1AT, which is abbreviated PI for this reason). A1AT is indeed the protease inhibitor most often involved in disease, namely in alpha 1-antitrypsin deficiency.
Protease inhibitors may be classified either by the type of protease they inhibit, or by their mechanism of action. In 2004 Rawlings and colleagues introduced a classification of protease inhibitors based on similarities detectable at the level of amino acid sequence. This classification initially identified 48 families of inhibitors that could be grouped into 26 related superfamily (or clans) by their structure. According to the MEROPS database there are now 85 families of inhibitors. These families are named with an I followed by a number, for example, I14 contains hirudin-like inhibitors.
Classes of proteases are:
Classes of inhibitor mechanisms of action are:
This is a family of protease suicide inhibitors called the serpins. It contains inhibitors of multiple cysteine and serine protease families. Their mechanism of action relies on undergoing a large conformational change which inactivates their target's catalytic triad.
Proteinase propeptide inhibitors (sometimes referred to as activation peptides) are responsible for the modulation of folding and activity of the peptidase pro-enzyme or zymogen. The pro-segment docks into the enzyme, shielding the substrate binding site, thereby promoting inhibition of the enzyme. Several such propeptides share a similar topology, despite often low sequence identities. The propeptide region has an open-sandwich antiparallel-alpha/antiparallel-beta fold, with two alpha-helices and four beta-strands with a (beta/alpha/beta)x2 topology. The peptidase inhibitor I9 family contains the propeptide domain at the N-terminus of peptidases belonging to MEROPS family S8A, subtilisins. The propeptide is removed by proteolytic cleavage; removal activating the enzyme.
...
Wikipedia




