PAS domain
| PAS fold | |||||||||
|---|---|---|---|---|---|---|---|---|---|
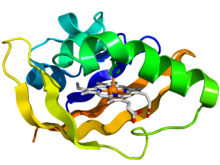
Crystallographic structure of the PAS domain of the bacterial oxygen sensor protein fixL. The protein is depicted as a rainbow colored cartoon (N-terminus = blue, C-terminus = red) while the heme ligand is shown as sticks (carbon = white, nitrogen = blue, oxygen = red, iron = orange).
|
|||||||||
| Identifiers | |||||||||
| Symbol | PAS | ||||||||
| Pfam | PF00989 | ||||||||
| InterPro | IPR013767 | ||||||||
| SMART | PAS | ||||||||
| PROSITE | PDOC50112 | ||||||||
| SCOP | 2phy | ||||||||
| SUPERFAMILY | 2phy | ||||||||
| CDD | cd00130 | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. Generally, the PAS domain acts as a molecular velcro, whereby small molecules and other proteins associate via binding of the PAS domain. Due to this velcro capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor.
PAS domains are found in a large number of organisms from bacteria to mammals. The PAS domain was named after the three proteins in which it was first discovered:
Per – period circadian protein
Arnt – aryl hydrocarbon receptor nuclear translocator protein
Sim – single-minded protein
Since the initial discovery of the PAS domain, a large quantity of PAS domain binding sites have been discovered in bacteria and eukaryotes. A subset called PAS LOV proteins are responsive to oxygen, light and voltage.
Although the PAS domain exhibits a degree of sequence variability, the three-dimensional structure of the PAS domain core is broadly conserved. This core consists of a five-stranded antiparallel β-sheet and several α-helices. Structural changes, as a result of signaling, predominantly originate within the β-sheet. These signals propagate via the α-helices of the core to the covalently-attached effector domain. In 1998, the PAS domain core architecture was first characterized in the structure of Halorhodospira halophila photoactive yellow protein (PYP). In many proteins, a dimer of PAS domains is required, whereby one binds a ligand and the other mediates interactions with other proteins.
The PAS domains that are known share less than 20% average pairwise sequence identity, meaning they are surprisingly dissimilar. PAS domains are frequently found on proteins with other environmental sensing mechanisms. Also, many PAS domains are attached to photoreceptive cells.
...
Wikipedia
