Mycolactone
 |
|
| Names | |
|---|---|
|
IUPAC name
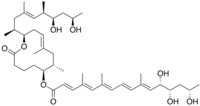
[(6S,7S,9E,12R)-12-[(E,2S,6R,7R,9R)-7,9-dihydroxy-4,6-dimethyldec-4-en-2-yl]-7,9-dimethyl-2-oxo-1-oxacyclododec-9-en-6-yl] (2E,4E,6E,8E,10E,12S,13S,15S)-12,13,15-trihydroxy-4,6,10-trimethylhexadeca-2,4,6,8,10-pentaenoate
|
|
| Identifiers | |
|
3D model (JSmol)
|
|
| ChemSpider | |
| MeSH | Mycolactone |
|
PubChem CID
|
|
|
|
|
|
| Properties | |
| C44H70O9 | |
| Molar mass | 743.021 |
|
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
|
|
|
|
|
| Infobox references | |
Mycolactone is a polyketide-derived macrolide produced and secreted by a group of very closely related pathogenic Mycobacteria species that have been assigned a variety of names including, M. ulcerans, M. liflandii (an unofficial designation), M. pseudoshottsii, and some strains of M. marinum. These mycobacteria are collectively referred to as mycolactone-producing mycobacteria or MPM.
In humans, mycolactone is the toxin responsible for Buruli ulcers, doing so by damaging tissues and inhibiting the immune response.
Five distinct, naturally occurring mycolactone structural variants have been described so far:
Mycolactone is produced by MPM through condensation of two polyketide chains (known as the core and acyl side chain). Different MPM produce characteristic mixtures of mycolactone congeners. The structural heterogeneity of mycolactones is due to variations in the acyl side chain. The structure of the mycolactone core is invariant.
The genes required for mycolactone biosynthesis form a contiguous 110-kb cluster on a large plasmid. The lactone core is produced by two polyketide synthases (PKS) that are encoded by the genes, mlsA1 and mlsA2, and a third polyketide synthase, encoded by the mlsB gene, produces the fatty acid-acyl side chain. Three putative accessory genes are found in the mycolactone cluster. One of these, MUP053, encodes a p450 monooxygenase that is thought to produce the hydroxyl at C′-12 on the fatty acid side chain. The gene encoding a FabH-like, type III ketosynthase (KS), located upstream of mlsA1, encodes a putative “joinase” (MUP045), and a type II thioesterase (TE II) gene (MUP038) is located between mlsA2 and mlsB.
Mycolactone consists of a 12-membered macrolide core with an ester-linked polyketide chain. Three plasmid-encoded polyketide synthase (PKS) enzymes are responsible for its production: MLSA 1 and MLSA 2 which generate the core, and MLSB is responsible for the synthesis of the polyketide chain. As shown in Figure 1, MLSB (1.2 MDa) contains seven consecutive extension modules and MLSA 1 (1.8 MDa) consists of eight. The remaining PKS enzyme, MLSA 2, contains the ninth module of MLSA. The C-terminal domains of both MLSA2 and MLSB includes a thioesterase (TE) that was thought to catalyze the formation of the mycolactone core but appears inactive. Each module consists of either malonyl-CoA or methylmalonyl-CoA Acyltransferase (AT) that allows for chain extension, a ketosynthase (KS), which catalyzes chain elongation, and an Acyl carrier protein (ACP) where the growing polyketide chain is attached. Modules may also consist of any of the following modifying domains: a dehydratase (DH), an enoyl reductase (ER) and one of two types of ketoreductase (KR) domains. Type A and B KRs refer to the two directions of ketoreduction that are correlated with specific amino acids in the active site. Four of the DH domains are predicted to be inactive based on a point mutation found in the active site sequence.
...
Wikipedia
