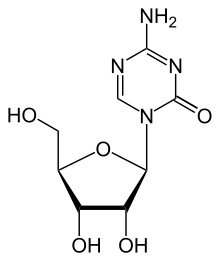
Azacitidine
 |
|
| Clinical data | |
|---|---|
| Trade names | Vidaza, Azadine |
| AHFS/Drugs.com | Monograph |
| MedlinePlus | a607068 |
| Pregnancy category |
|
| Routes of administration |
SubQ, IV |
| ATC code | |
| Legal status | |
| Legal status |
|
| Pharmacokinetic data | |
| Metabolism | possible hepatic metabolism, mostly urinary excretion |
| Biological half-life | 4 hr. |
| Identifiers | |
|
|
| Synonyms | 5-azacytidine, Azacytidine, 320-67-2; Ladakamycin, 4-Amino-1-beta-D-ribofuranosyl-s-triazin-2(1H)-one, U-18496 |
| CAS Number | |
| PubChem CID | |
| IUPHAR/BPS | |
| DrugBank | |
| ChemSpider | |
| UNII | |
| KEGG | |
| ChEBI | |
| ChEMBL | |
| ECHA InfoCard | 100.005.711 |
| Chemical and physical data | |
| Formula | C8H12N4O5 |
| Molar mass | 244.205 g/mol |
| 3D model (Jmol) | |
|
|
|
|
Azacitidine (INN; trade name Vidaza) is a chemical analog of cytidine, a nucleoside in DNA and RNA. Azacitidine and its deoxy derivative, decitabine (also known as 5-aza-2′deoxycytidine), are used in the treatment of myelodysplastic syndrome. Both drugs were first synthesized in Czechoslovakia as potential chemotherapeutic agents for cancer.
Azacitidine, marketed as Vidaza, is used mainly in the treatment of myelodysplastic syndrome, for which it received approval by the U.S. Food and Drug Administration on May 19, 2004. In two randomized controlled trials comparing azacitidine to supportive treatment, 16% of subjects with myelodysplastic syndrome who were randomized to receive azacitidine had a complete or partial normalization of blood cell counts and bone marrow morphology, compared to none who received supportive care, and about two-thirds of patients who required blood transfusions no longer needed them after receiving azacitidine.
It is also sometimes used for the treatment of acute myeloid leukemia, as a hypomethylating agent; an oral version called CC-486 is being tested as an easier to administer treatment for AML.
Azacitidine can be used in vitro to remove methyl groups from DNA. This may weaken the effects of gene silencing mechanisms that occur prior to methylation. Certain methylations are believed to secure DNA in a silenced state, and therefore demethylation may reduce the stability of silencing signals and confer relative gene activation.
...
Wikipedia
