U4 spliceosomal RNA
| U4 spliceosomal RNA | |
|---|---|
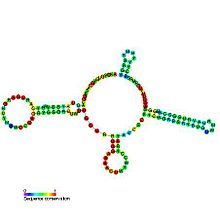
Predicted secondary structure and sequence conservation of U4
|
|
| Identifiers | |
| Symbol | U4 |
| Rfam | RF00015 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0017070 0000353 0000351 0005687 0046540 |
| SO | 0000393 |
The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.
The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNP, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been proposed to coincide with different temporal events in the activity of the penta-snRNP, or as intermediates in the step-wise model of spliceosome assembly and activity.
The U4 snRNA (and its likely analog snR14 in Yeast) has been shown not to participate directly in the specific catalytic activities of the splicing reaction, and is proposed instead to act as a regulator of the U6 snRNA. The U4 snRNA inhibits spliceosome activity during assembly by complementary base pairing between the U6 snRNA in two highly conserved stem regions. It is suggested that this base-pairing interaction prevents the U6 snRNA from assembling with the U2 snRNA into the conformation required for catalytic activity. If the U4 snRNA is degraded and thereby removed from the spliceosome, splicing is effectively halted. The U4 and U6 snRNAs are demonstratively required for splicing in vitro.
The U4 snRNA secondary structure is suggested to alter depending on its interaction with the U6 snRNA. Several experiments involving X-ray crystallography,NMR, and chemical modification RNA structure probing indicate that U4 snRNA secondary structure contains several conserved motifs, which serve structural as well as intermediary roles in establishing interactions with other splicing components. The putative U4/U6 snRNA base pairing secondary structure shown in Figure 2., is conserved across a diverse set of organisms suggesting the splicing machinery's ancient origins. It has been shown previously that a highly conserved Kinked-loop participates in specific protein interactions.
...
Wikipedia
