Self-splicing intron
| Group I catalytic intron | |
|---|---|
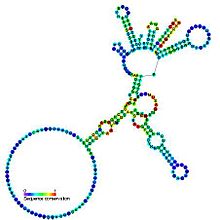
Predicted secondary structure and sequence conservation of Group I catalytic intron
|
|
| Identifiers | |
| Symbol | Intron_gpI |
| Rfam | RF00028 |
| Other data | |
| RNA type | Intron |
| Domain(s) | Eukaryota; Bacteria; Viruses |
| GO | 0000372 |
| SO | 0000587 |
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains - the P4-P6 domain (formed from the stacking of P5, P4, P6 and P6a helices) and the P3-P9 domain (formed from the P8, P3, P7 and P9 helices). The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.
Splicing of group I introns is processed by two sequential ester-transfer reactions. The exogenous guanosine or guanosine nucleotide (exoG) first docks onto the active G-binding site located in P7, and its 3'-OH is aligned to attack the phosphodiester bond at the 5' splice site located in P1, resulting in a free 3'-OH group at the upstream exon and the exoG being attached to the 5' end of the intron. Then the terminal G (omega G) of the intron swaps the exoG and occupies the G-binding site to organize the second ester-transfer reaction: the 3'-OH group of the upstream exon in P1 is aligned to attack the 3' splice site in P10, leading to the ligation of the adjacent upstream and downstream exons and release of the catalytic intron.
Two-metal-ion mechanism seen in protein polymerases and phosphatases was proposed to be used by group I and group II introns to process the phosphoryl transfer reactions, which was unambiguously proven by a recently resolved high-resolution structure of the Azoarcus group I intron.
...
Wikipedia
