Biliverdin reductase
| biliverdin reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 |
|||||||||
| Identifiers | |||||||||
| EC number | 1.3.1.24 | ||||||||
| CAS number | 9074-10-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
|
|||||||||
| Search | |
|---|---|
| PMC | articles |
| PubMed | articles |
| NCBI | proteins |
| biliverdin reductase A | |
|---|---|
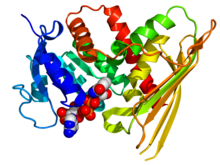
Crystallographic structure of human biliverdin reductase A based on the PDB: 2H63 coordinates. The enzyme is displayed as a rainbow colored cartoon (N-terminus = blue, C-terminus = red) while the NADP cofactor is displayed as space-filling model (carbon = white, oxygen = red, nitrogen = blue, phosphorus = orange).
|
|
| Identifiers | |
| Symbol | BLVRA |
| Alt. symbols | BLVR |
| Entrez | 644 |
| HUGO | 1062 |
| OMIM | 109750 |
| RefSeq | NM_000712 |
| UniProt | P53004 |
| Other data | |
| EC number | 1.3.1.24 |
| Locus | Chr. 7 p14-cen |
| biliverdin reductase B | |
|---|---|
| Identifiers | |
| Symbol | BLVRB |
| Alt. symbols | FLR |
| Entrez | 645 |
| HUGO | 1063 |
| OMIM | 600941 |
| RefSeq | NM_000713 |
| UniProt | P30043 |
| Other data | |
| EC number | 1.3.1.24 |
| Locus | Chr. 19 q13.1-13.2 |
| Biliverdin reductase, catalytic | |||||||||
|---|---|---|---|---|---|---|---|---|---|

crystal structure of a biliverdin reductase enzyme-cofactor complex
|
|||||||||
| Identifiers | |||||||||
| Symbol | Biliv-reduc_cat | ||||||||
| Pfam | PF09166 | ||||||||
| InterPro | IPR015249 | ||||||||
| SCOP | 1lc0 | ||||||||
| SUPERFAMILY | 1lc0 | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
Biliverdin reductase (BVR) is an enzyme (EC 1.3.1.24) found in all tissues under normal conditions, but especially in reticulo-macrophages of the liver and spleen. BVR facilitates the conversion of biliverdin to bilirubin via the reduction of a double-bond between the second and third pyrrole ring into a single-bond.
There are two isozymes, in humans, each encoded by its own gene, biliverdin reductase A (BLVRA) and biliverdin reductase B (BLVRB).
BVR acts on biliverdin by reducing its double-bond between the pyrrole rings into a single-bond. It accomplishes this using NADPH + H+ as an electron donor, forming bilirubin and NADP+ as products.
BVR catalyzes this reaction through an overlapping binding site including Lys18, Lys22, Lys179, Arg183, and Arg185 as key residues. This binding site attaches to biliverdin, and causes its dissociation from heme oxygenase (HO) (which catalyzes reaction of ferric heme --> biliverdin), causing the subsequent reduction to bilirubin.
BVR is composed of two closely packed domains, between 247-415 amino acids long and containing a Rossmann fold. BVR has also been determined to be a zinc-binding protein with each enzyme protein having one strong-binding zinc atom.
The C-terminal half of BVR contains the catalytic domain, which adopts a structure containing a six-stranded beta-sheet that is flanked on one face by several alpha-helices. This domain contains the catalytic active site, which reduces the gamma-methene bridge of the open tetrapyrrole, biliverdin IX alpha, to bilirubin with the concomitant oxidation of a NADH or NADPH cofactor.
...
Wikipedia
