Aconitase
| aconitate hydratase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
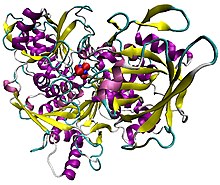
Illustration of pig aconitase in complex with the [Fe4S4] cluster. The protein is colored by secondary structure, and iron atoms are blue and the sulfur red.
|
|||||||||
| Identifiers | |||||||||
| EC number | 4.2.1.3 | ||||||||
| CAS number | 9024-25-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
|
|||||||||
| Search | |
|---|---|
| PMC | articles |
| PubMed | articles |
| NCBI | proteins |
| Aconitase family (aconitate hydratase) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|

Structure of aconitase.
|
|||||||||
| Identifiers | |||||||||
| Symbol | Aconitase | ||||||||
| Pfam | PF00330 | ||||||||
| InterPro | IPR001030 | ||||||||
| PROSITE | PDOC00423 | ||||||||
| SCOP | 1aco | ||||||||
| SUPERFAMILY | 1aco | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
| aconitase 1, soluble | |
|---|---|
| Identifiers | |
| Symbol | ACO1 |
| Alt. symbols | IREB1 |
| Entrez | 48 |
| HUGO | 117 |
| OMIM | 100880 |
| RefSeq | NM_002197 |
| UniProt | P21399 |
| Other data | |
| EC number | 4.2.1.3 |
| Locus | Chr. 9 p21.1 |
| aconitase 2, mitochondrial | |
|---|---|
| Identifiers | |
| Symbol | ACO2 |
| Alt. symbols | ACONM |
| Entrez | 50 |
| HUGO | 118 |
| OMIM | 100850 |
| RefSeq | NM_001098 |
| UniProt | Q99798 |
| Other data | |
| EC number | 4.2.1.3 |
| Locus | Chr. 22 q13.2 |
Aconitase (aconitate hydratase; EC 4.2.1.3) is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.
Aconitase, displayed in the structures in the right margin of this page, has two slightly different structures, depending on whether it is activated or inactivated. In the inactive form, its structure is divided into four domains. Counting from the N-terminus, only the first three of these domains are involved in close interactions with the [3Fe-4S] cluster, but the active site consists of residues from all four domains, including the larger C-terminal domain. The Fe-S cluster and a SO42− anion also reside in the active site. When the enzyme is activated, it gains an additional iron atom, creating a [4Fe-4S] cluster. However, the structure of the rest of the enzyme is nearly unchanged; the conserved atoms between the two forms are in essentially the same positions, up to a difference of 0.1 angstroms.
In contrast with the majority of iron-sulfur proteins that function as electron carriers, the iron-sulfur cluster of aconitase reacts directly with an enzyme substrate. Aconitase has an active [Fe4S4]2+ cluster, which may convert to an inactive [Fe3S4]+ form. Three cysteine (Cys) residues have been shown to be ligands of the [Fe4S4] centre. In the active state, the labile iron ion of the [Fe4S4] cluster is not coordinated by Cys but by water molecules.
The iron-responsive element-binding protein (IRE-BP) and 3-isopropylmalate dehydratase (α-isopropylmalate isomerase; EC 4.2.1.33), an enzyme catalysing the second step in the biosynthesis of leucine, are known aconitase homologues. Iron regulatory elements (IREs) constitute a family of 28-nucleotide, non-coding, stem-loop structures that regulate iron storage, heme synthesis and iron uptake. They also participate in ribosome binding and control the mRNA turnover (degradation). The specific regulator protein, the IRE-BP, binds to IREs in both 5' and 3' regions, but only to RNA in the apo form, without the Fe-S cluster. Expression of IRE-BP in cultured cells has revealed that the protein functions either as an active aconitase, when cells are iron-replete, or as an active RNA-binding protein, when cells are iron-depleted. Mutant IRE-BPs, in which any or all of the three Cys residues involved in Fe-S formation are replaced by serine, have no aconitase activity, but retain RNA-binding properties.
...
Wikipedia
