Crossover (genetic algorithm)
In genetic algorithms, crossover is a genetic operator used to vary the programming of a chromosome or chromosomes from one generation to the next. It is analogous to reproduction and biological crossover, upon which genetic algorithms are based. Cross over is a process of taking more than one parent solutions and producing a child solution from them. There are methods for selection of the chromosomes. Those are also given below.
Many crossover techniques exist for organisms which use different data structures to store themselves.
A single crossover point on both parents' organism strings is selected. All data beyond that point in either organism string is swapped between the two parent organisms. The resulting organisms are the children:
Two-point crossover calls for two points to be selected on the parent organism strings. Everything between the two points is swapped between the parent organisms, rendering two child organisms:
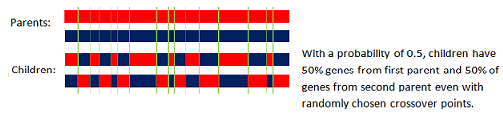
The uniform crossover uses a fixed mixing ratio between two parents. Unlike single- and two-point crossover, the uniform crossover enables the parent chromosomes to contribute the gene level rather than the segment level.
If the mixing ratio is 0.5, the offspring has approximately half of the genes from first parent and the other half from second parent, although cross over points can be randomly chosen as seen below:
...
Wikipedia