Histones
| Core histone H2A/H2B/H3/H4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
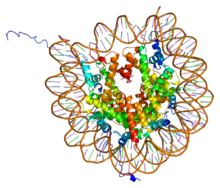
PDB rendering of Complex between nucleosome core particle (h3,h4,h2a,h2b) and 146 bp long DNA fragment based on 1aoi.
|
|||||||||
| Identifiers | |||||||||
| Symbol | Histone | ||||||||
| Pfam | PF00125 | ||||||||
| Pfam clan | CL0012 | ||||||||
| InterPro | IPR007125 | ||||||||
| SCOP | 1hio | ||||||||
| SUPERFAMILY | 1hio | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
| linker histone H1 and H5 family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|||||||||
| Identifiers | |||||||||
| Symbol | Linker_histone | ||||||||
| Pfam | PF00538 | ||||||||
| InterPro | IPR005818 | ||||||||
| SMART | SM00526 | ||||||||
| SCOP | 1hst | ||||||||
| SUPERFAMILY | 1hst | ||||||||
|
|||||||||
| Available protein structures: | |
|---|---|
| Pfam | structures |
| PDB | RCSB PDB; PDBe; PDBj |
| PDBsum | structure summary |
In biology, histones are highly alkaline proteins found in eukaryotic cell nuclei that package and order the DNA into structural units called nucleosomes. They are the chief protein components of chromatin, acting as spools around which DNA winds, and playing a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to 1 in human DNA). For example, each human diploid cell (containing 23 pairs of chromosomes) has about 1.8 meters of DNA, but wound on the histones it has about 90 micrometers (0.09 mm) of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes.
Five major families of histones exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain; three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif). The resulting four distinct dimers then come together to form one octameric nucleosome core, approximately 63 Angstroms in diameter (a solenoid (DNA)-like particle). Around 146 base pairs (bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across. The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA separating each pair of nucleosomes (also referred to as linker DNA). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
...
Wikipedia
